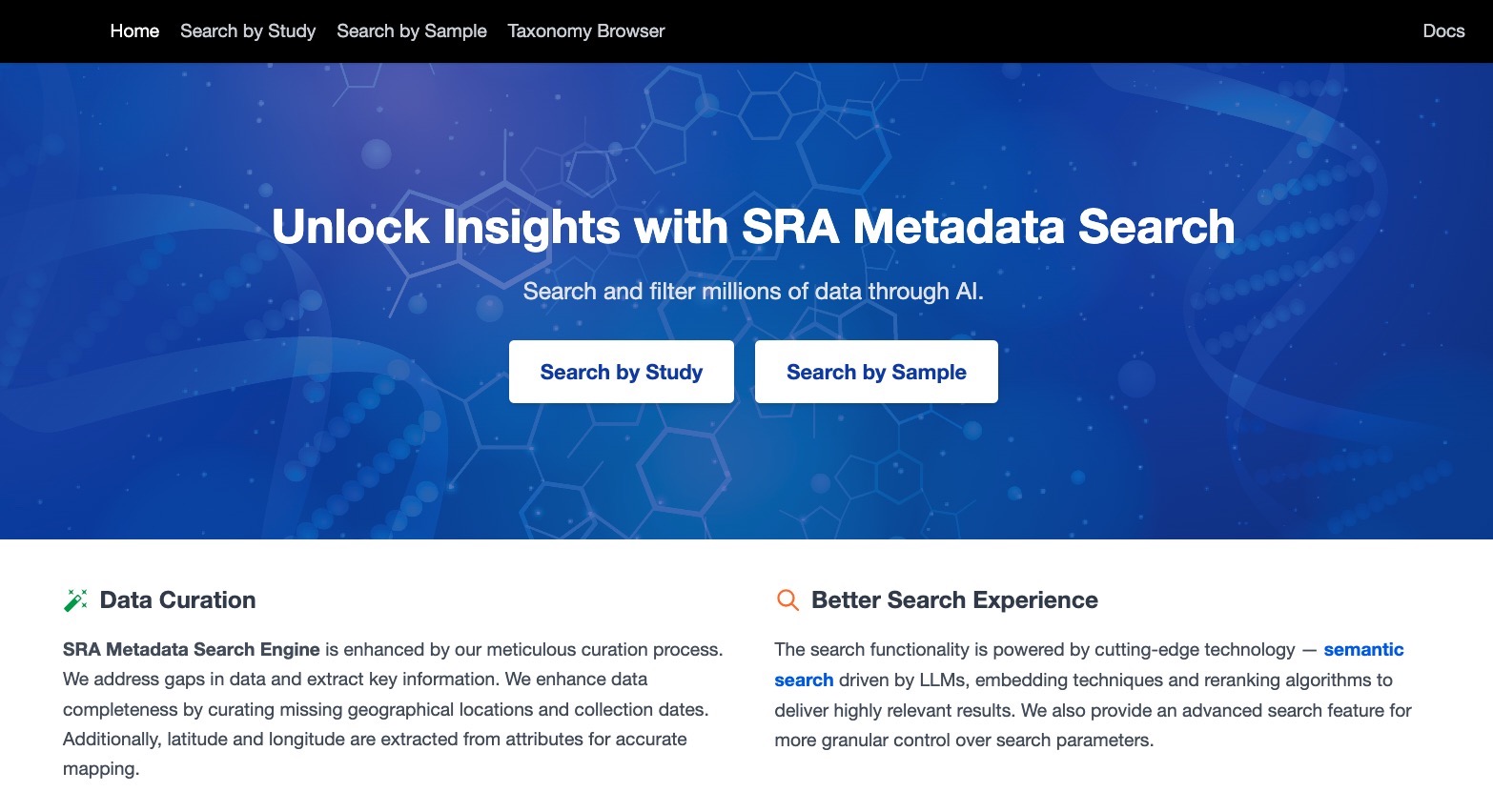
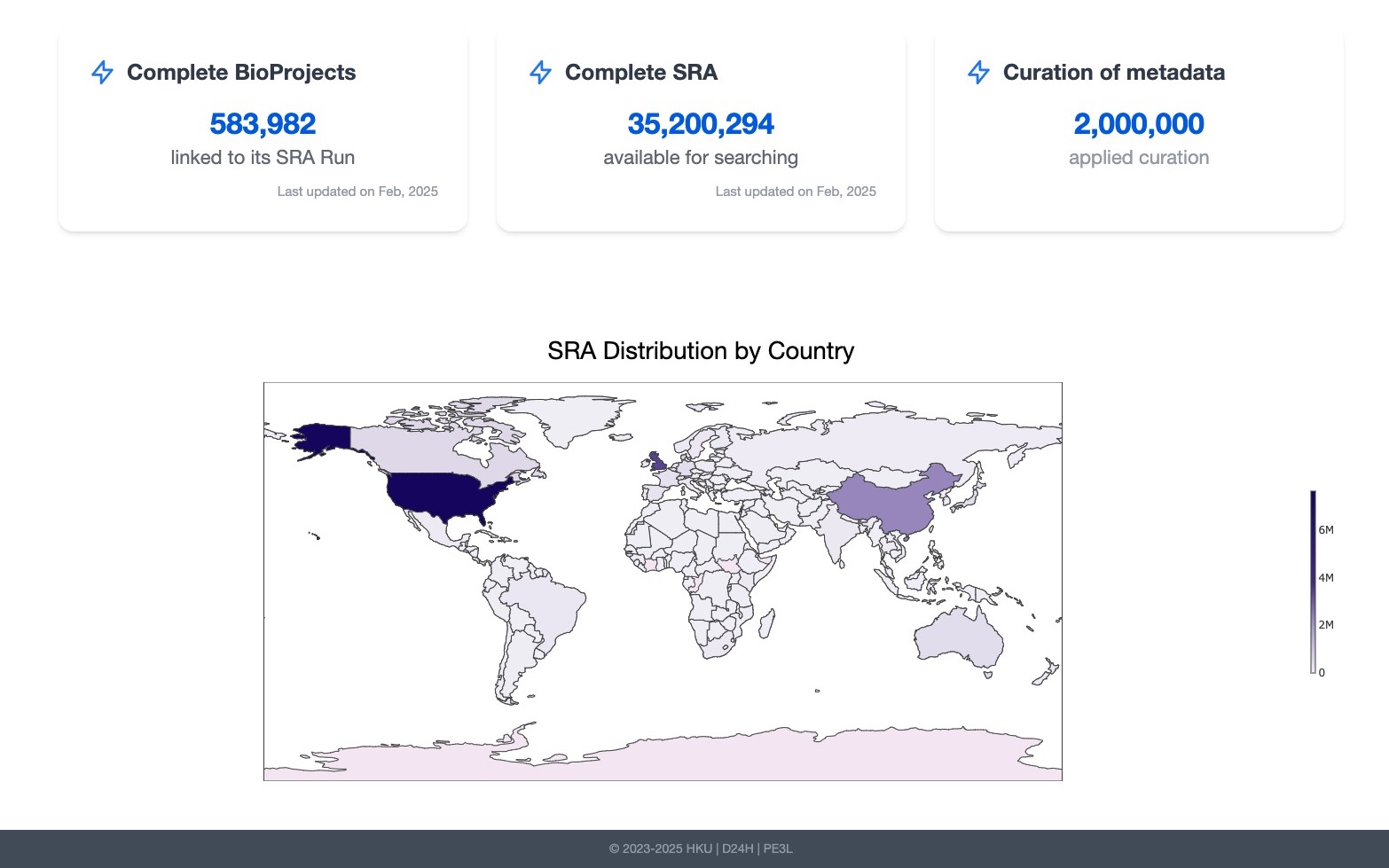
LLM-powered Semantic Search Platform
🧬 SRA Metadata Search Tool
Role: AI Programmer
Company: D24H
Project Period: Jan 2025 – Jun 2025
“Developed a production-grade, open-source, natural language search engine for the world’s largest public genomic resource — the Sequence Read Archive (SRA) — enabling researchers to query and filter 35M+ records using semantic search, advanced filters, and state-of-the-art AI models.”
Project Highlights
- LLM Integration: Built an end-to-end query interpretation pipeline using Gemini-2.0-flash-lite for natural language parsing, typo correction, and metadata extraction.
- Semantic & ANN Search: Engineered fast, accurate semantic search with BERT-based SentenceTransformers and pgvector, supporting <0.2s query speed on 35M+ samples.
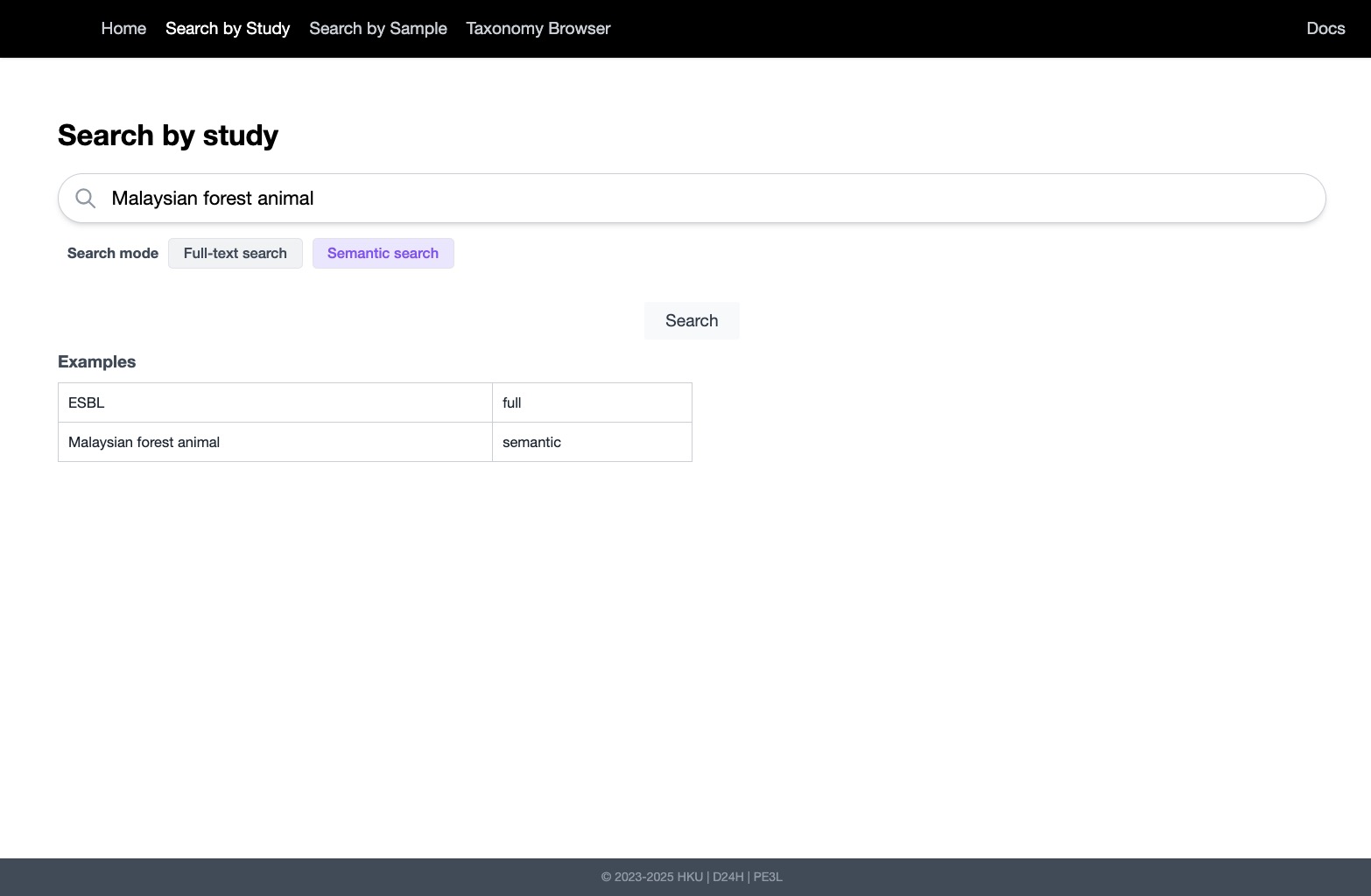
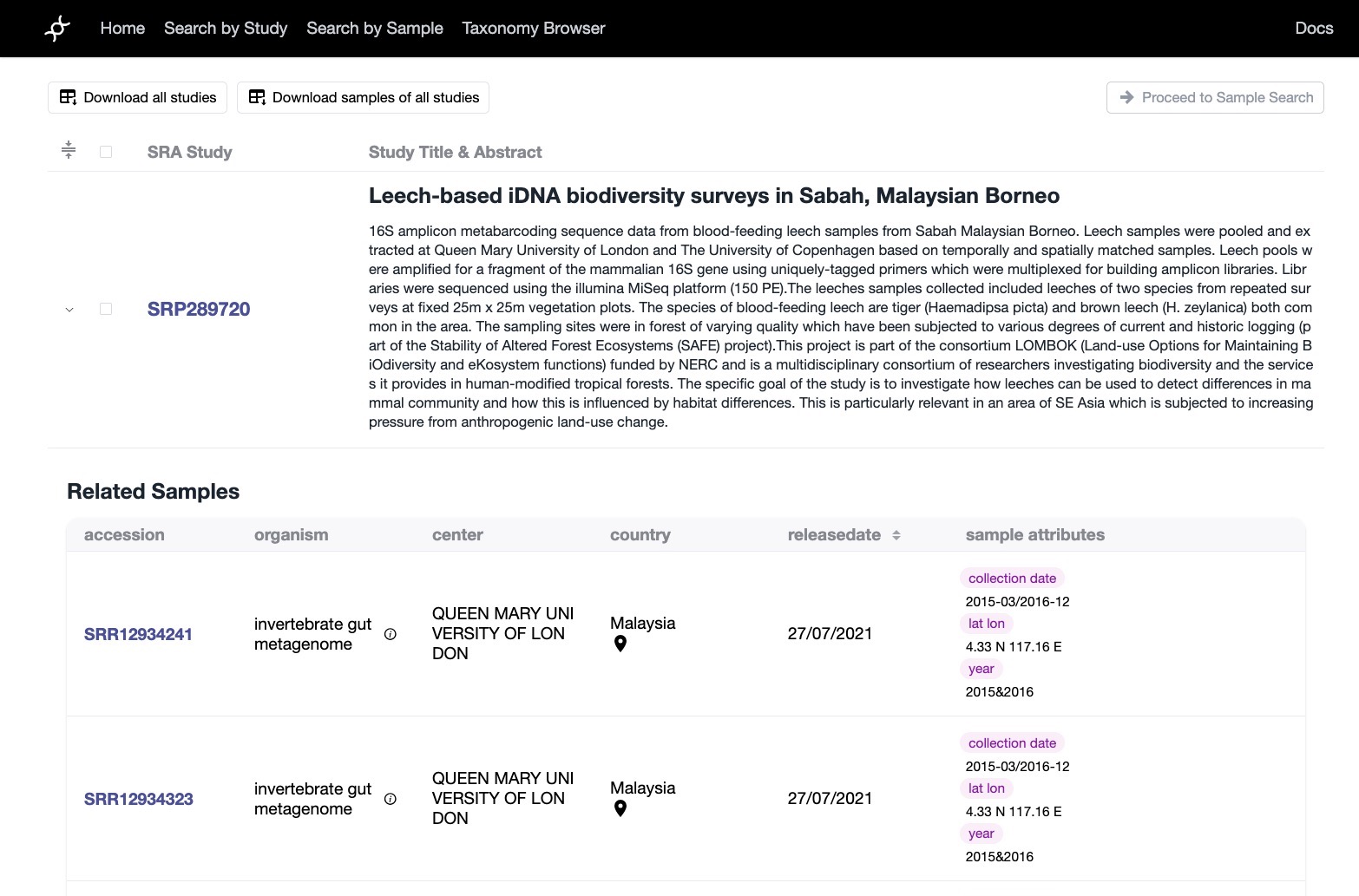
- Advanced Filtering: Supported complex queries on organism, geography, time, sequencing platform, and more, combining both semantic and structured filters.
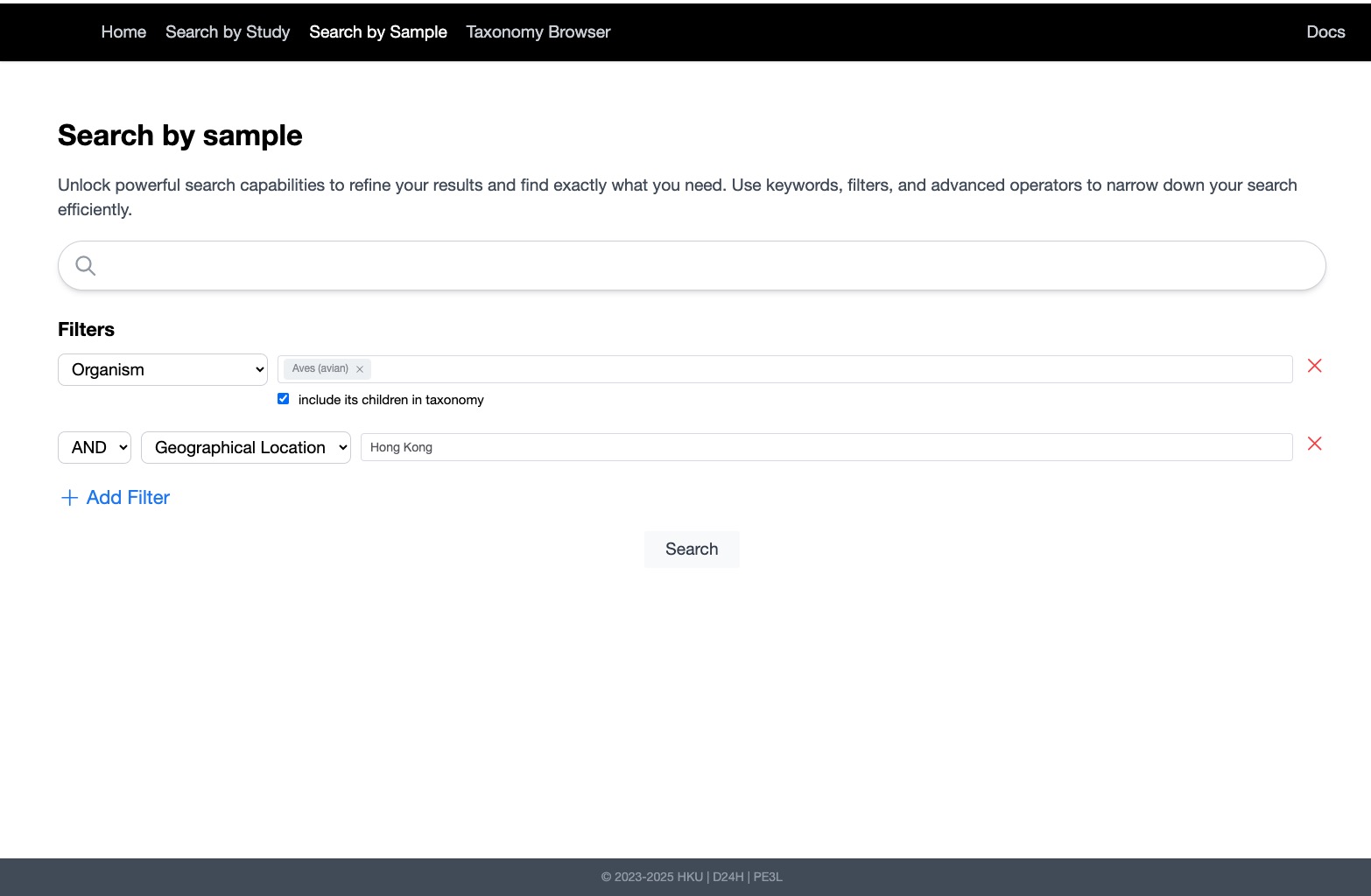
- Reranking & Relevance: Integrated transformer-based cross-encoder models (Jina AI) for context-aware reranking of search results.
- User Interface: Designed a researcher-friendly UI for flexible searching and immediate data exploration, removing the need for SQL or bioinformatics expertise.
- Scalable Infrastructure: Leveraged PostgreSQL 16, pgvector 0.8.0, and efficient embedding storage to achieve high throughput and low-cost querying at scale.
Key Technologies: Python, Flask, PostgreSQL, pgvector, SentenceTransformers, JinaAI, Pandas, Torch, LLMs